MADS-box genes and microProteins:
implications for transcription factor evolution and regulation
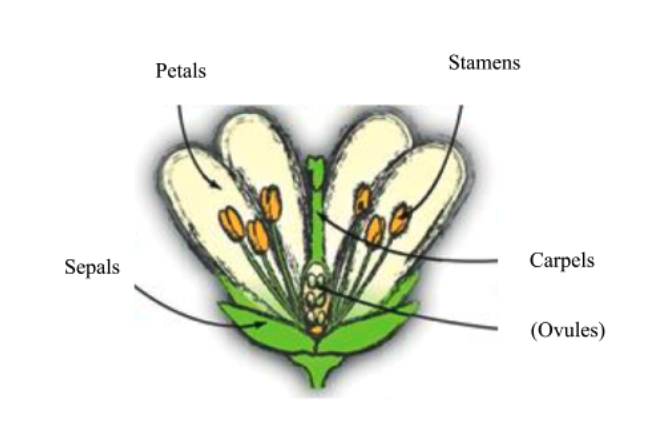
Across flowering plants, floral structure is variant, but most are generally composed of a combination of sepals, petals, stamens, and carpels, the identity of which are specified by MADS-box transcription factor genes.
The highly-conserved floral homeotic MADS-box genes of the MIKC-type MADS subfamily plan important roles in agriculture as disruption to protein function leads to phenotypic consequences in flower and fruit size. Transcription factors, like the MADS-box family, may be regulated by other factors such as microProteins. MicroProteins (miPs) are small proteins that typically consist of a single protein-protein interaction domain.
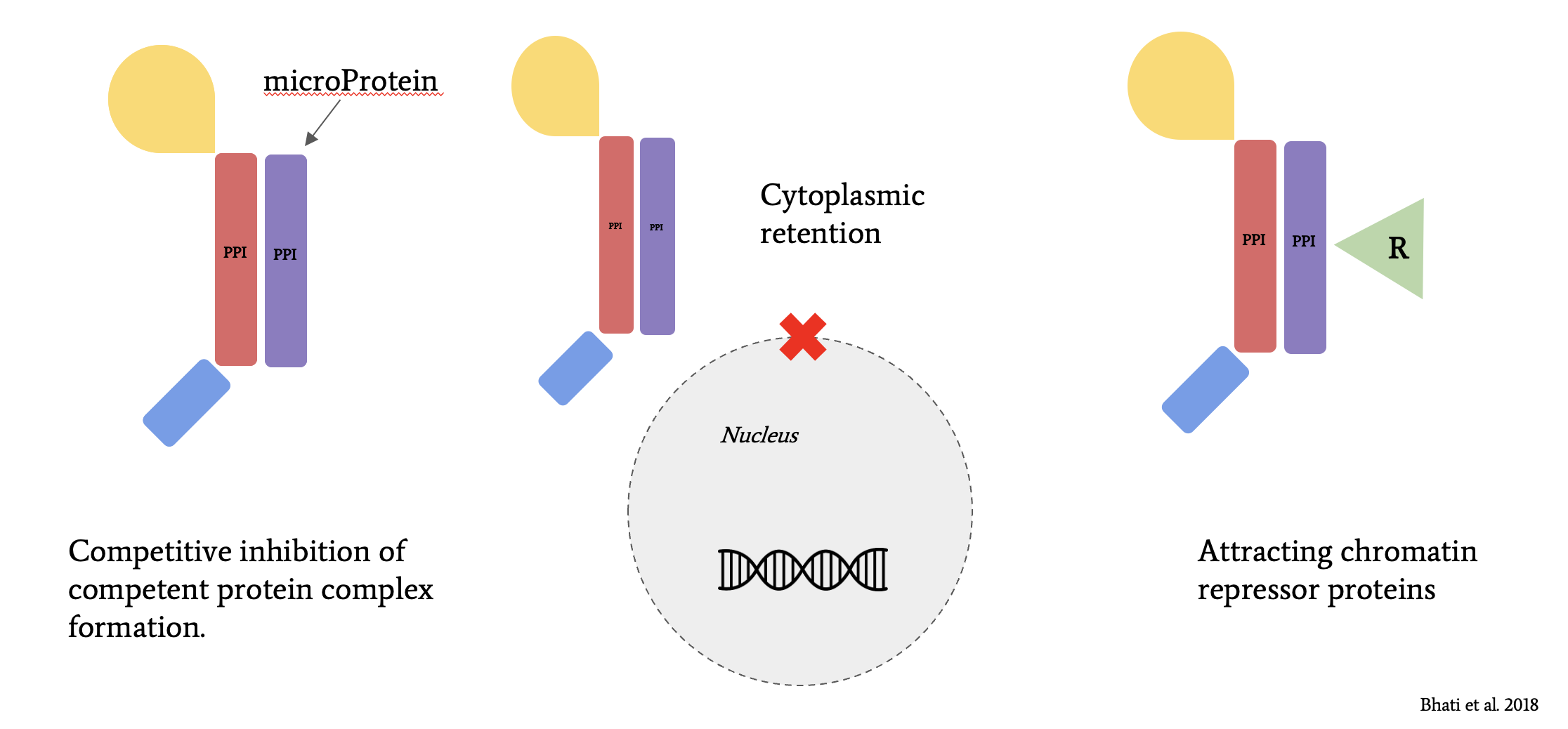
These families of small proteins act as negative regulators as their expression leads to repression of their related target proteins. While several subfamilies of miPs have been classified according to the family of transcription factors with which they interact, no previous literature exists identifying miPs that regulate MADS-box family.
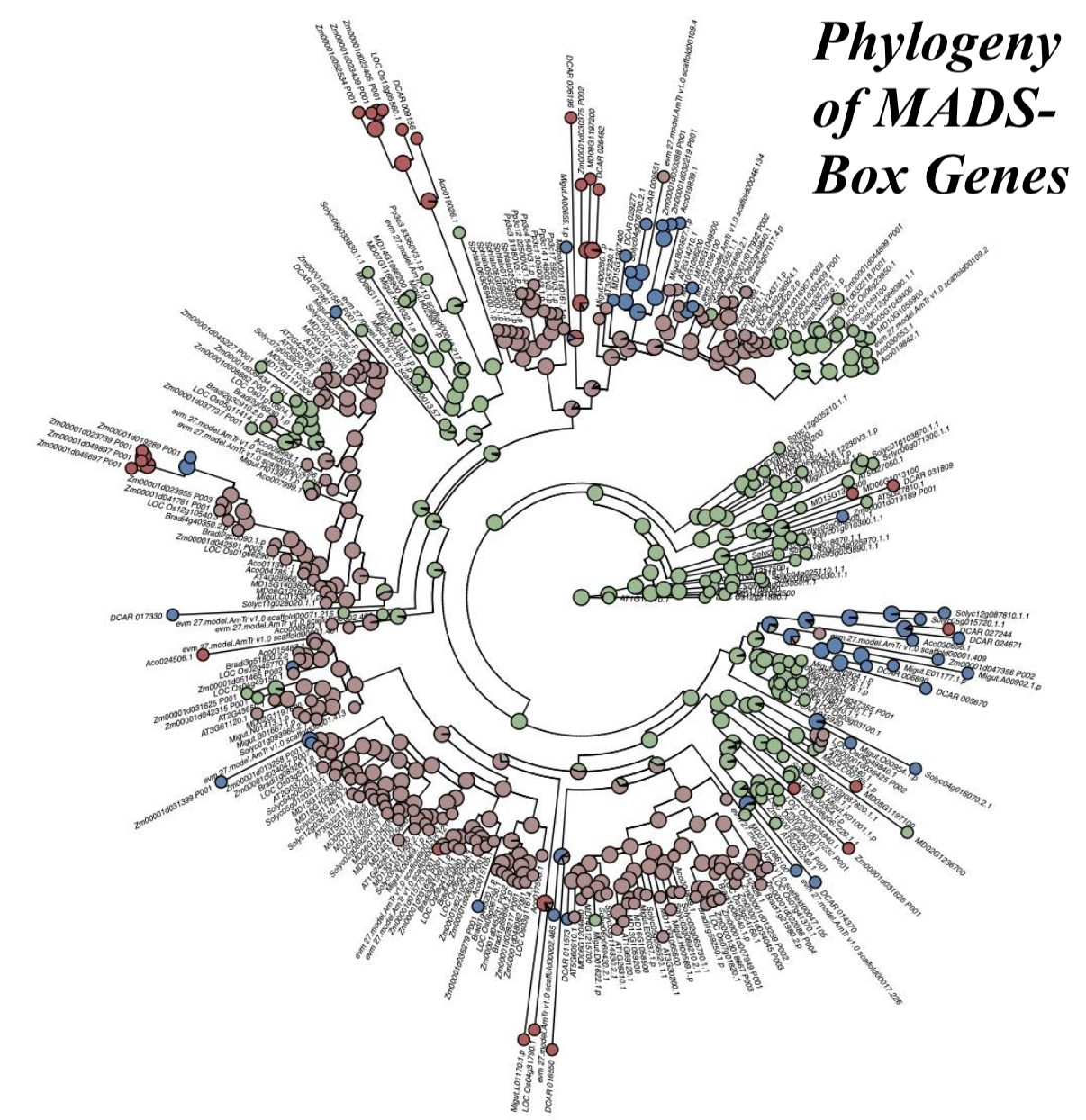
In order to determine whether microProteins may exist within the MADS-box gene family, I performed searches for poteintial miPs using Hidden Markov Model (HMM) profile searching and protein domain profiling. From these profiles and subsequent gene alignments, I identified 10 strong candidate miPs within the MADS-box gene family that have strong consensus in the protein-protein interaction domains of full-length MADS-box genes.
Advisors: Dr. Madelaine Bartlett, Dr. Jarrett Man
University of Massachusetts, Commonwealth Honors College
![]()